Bootstrapping a vector of results, by group in R
Question: How can I use a boostrap to get confidence intervals for a collection of statistics calculated on the eigenvalues of covariance matrices, separately for each group (factor level) in a data frame?
Problem: I can't quite work out the data
structure I need to contain these results suitable for the boot function, or a way to "map" the bootstrap over groups and obtain confidence intervals in a form suitable for plotting.
Context:
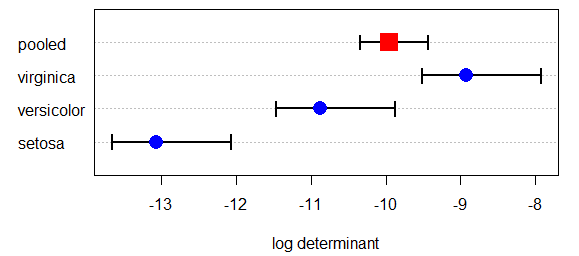
In the heplots package, boxM calculates Box's M test of equality of covariance matrices.
There is a plot method that produces a useful plot of the log determinants that go into this
test. The confidence intervals in this plot are based on an asymptotic theory approximation.
> library(heplots)
> iris.boxm <- boxM(iris[, 1:4], iris[, "Species"])
> iris.boxm
Box's M-test for Homogeneity of Covariance Matrices
data: iris[, 1:4]
Chi-Sq (approx.) = 140.94, df = 20, p-value < 2.2e-16
> plot(iris.boxm, gplabel="Species")
The plot method can also display other functions of the eigenvalues, but no theoretical confidence intervals are available in this case.
op <- par(mfrow=c(2,2), mar=c(5,4,1,1))
plot(iris.boxm, gplabel="Species", which="product")
plot(iris.boxm, gplabel="Species", which="sum")
plot(iris.boxm, gplabel="Species", which="precision")
plot(iris.boxm, gplabel="Species", which="max")
par(op)
Thus, I would like to be able to calculate these CIs using a boostrap, and display them in the corresponding plots.
What I've tried:
Below are functions that boostrap these statistics, but for the total
sample, not taking group (Species) into account.
cov_stat_fun <- function(data, indices,
stats=c("logdet", "prod", "sum", "precision", "max")
) {
dat <- data[indices,]
cov <- cov(dat, use="complete.obs")
eigs <- eigen(cov)$values
res <- c(
"logdet" = log(det(cov)),
"prod" = prod(eigs),
"sum" = sum(eigs),
"precision" = 1/ sum(1/eigs),
"max" = max(eigs)
)
}
boot_cov_stat <- function(data, R=500, ...) {
boot(data, cov_stat_fun, R=R, ...)
}
This works, but I need the results by group (and also for the total sample)
> iris.boot <- boot_cov_stat(iris[,1:4])
>
> iris.ci <- boot.ci(iris.boot)
> iris.ci
BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS
Based on 500 bootstrap replicates
CALL :
boot.ci(boot.out = iris.boot)
Intervals :
Level Normal Basic Studentized
95% (-6.622, -5.702 ) (-6.593, -5.653 ) (-6.542, -5.438 )
Level Percentile BCa
95% (-6.865, -5.926 ) (-6.613, -5.678 )
Calculations and Intervals on Original Scale
Some BCa intervals may be unstable
>
I also have written a function that calculates the separate covariance matrices for each group, but I can't see how to use this in my bootstrap functions. Can someone help?
# calculate covariance matrices by group and pooled
covs <- function(Y, group) {
Y <- as.matrix(Y)
gname <- deparse(substitute(group))
if (!is.factor(group)) group <- as.factor(as.character(group))
valid <- complete.cases(Y, group)
if (nrow(Y) > sum(valid))
warning(paste(nrow(Y) - sum(valid)), " cases with missing data have been removed.")
Y <- Y[valid,]
group <- group[valid]
nlev <- nlevels(group)
lev <- levels(group)
mats <- aux <- list()
for(i in 1:nlev) {
mats[[i]] <- cov(Y[group == lev[i], ])
}
names(mats) <- lev
pooled <- cov(Y)
c(mats, "pooled"=pooled)
}
Edit:
In a seemingly related question, Bootstrap by groups, it is suggested that an answer is provided by using the strata argument to boot(), but there is no example of what this gives. [Ah: the strata argument just assures that strata are represented in the bootstrap sample in relation to their frequencies in the data.]
Trying this for my problem, I am not further enlightened, because what I want to get is separate confidence intervals for each Species.
> iris.boot.strat <- boot_cov_stat(iris[,1:4], strata=iris$Species)
>
> boot.ci(iris.boot.strat, conf=0.95, type=c("basic", "bca"))
BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS
Based on 500 bootstrap replicates
CALL :
boot.ci(boot.out = iris.boot.strat, conf = 0.95, type = c("basic",
"bca"))
Intervals :
Level Basic BCa
95% (-6.587, -5.743 ) (-6.559, -5.841 )
Calculations and Intervals on Original Scale
Some BCa intervals may be unstable
>
If I understand your question, you can run your bootstrap function by group as follows:
library(boot)
library(tidyverse)
# Pooled
iris.boot <- boot_cov_stat(iris[,1:4])
iris.ci <- boot.ci(iris.boot)
# By Species
boot.list = setNames(unique(iris$Species), unique(iris$Species)) %>%
map(function(group) {
iris.boot = boot_cov_stat(iris[iris$Species==group, 1:4])
boot.ci(iris.boot)
})
# Combine pooled and by-Species results
boot.list = c(boot.list, list(Pooled=iris.ci))
boot.list
$setosa BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS Based on 500 bootstrap replicates CALL : boot.ci(boot.out = iris.boot) Intervals : Level Normal Basic Studentized 95% (-13.69, -11.86 ) (-13.69, -11.79 ) (-13.52, -10.65 ) Level Percentile BCa 95% (-14.34, -12.44 ) (-13.65, -11.99 ) Calculations and Intervals on Original Scale Warning : BCa Intervals used Extreme Quantiles Some BCa intervals may be unstable $versicolor BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS Based on 500 bootstrap replicates CALL : boot.ci(boot.out = iris.boot) Intervals : Level Normal Basic Studentized 95% (-11.37, -9.81 ) (-11.36, -9.78 ) (-11.25, -8.97 ) Level Percentile BCa 95% (-11.97, -10.39 ) (-11.35, -10.09 ) Calculations and Intervals on Original Scale Warning : BCa Intervals used Extreme Quantiles Some BCa intervals may be unstable $virginica BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS Based on 500 bootstrap replicates CALL : boot.ci(boot.out = iris.boot) Intervals : Level Normal Basic Studentized 95% (-9.467, -7.784 ) (-9.447, -7.804 ) (-9.328, -6.959 ) Level Percentile BCa 95% (-10.050, -8.407 ) ( -9.456, -8.075 ) Calculations and Intervals on Original Scale Warning : BCa Intervals used Extreme Quantiles Some BCa intervals may be unstable $Pooled BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS Based on 500 bootstrap replicates CALL : boot.ci(boot.out = iris.boot) Intervals : Level Normal Basic Studentized 95% (-6.620, -5.714 ) (-6.613, -5.715 ) (-6.556, -5.545 ) Level Percentile BCa 95% (-6.803, -5.906 ) (-6.624, -5.779 ) Calculations and Intervals on Original Scale Some BCa intervals may be unstable
- How to find correct projection of the shapefile data for leaflet map in r?
- How to use regex in R to shorten strings by one space
- R CRAN Check fail when using parallel functions
- Replacing several rows of data in a column efficiently using condition in a pipeline
- Iterate over vectors from an imported dataframe row-wise
- Split the legend into three rows with headings for a nested facet_wrap
- Why replace works with a boolean mask?
- Not able properly rasterize polygon dataset using terra::rasterize()
- Less than symbol ("<") in cloze schoice in Canvas (html select dropdown) shows as &lt;
- R/exams docx output does not include images
- How to have primitive discard additional arguments
- Functions Causing Objects to Disappear
- How to repel texts equidistantly on a secondary y-axis of a facet_grid?
- How can I add some text to a ggplot2 graph in R outside the plot area?
- Extracting Exact p-values from the "did" Package Output in R
- How can I set column types of several columns in empty R data frame?
- Adjust vertical line based on maximum value in graph plotly
- Combining Two Dataframes Based on Multiple Conditions & Removing Rows that Don't Match
- independent sample t-test in R using test statistics (mean, std. dev, count)
- How do I load a package without installing it in R?
- survcheck: 'x' must be an array of at least two dimensions
- Insert variable names and their specific values between the last column of facets and the strips of a facet_grid in R
- Use a recorded plot for officer
- Errors when using RStudio's Git tools
- Orientation of an irregular window changes the distance r over which secondary summary statistics are estimated
- Elegant way to check for missing packages and install them?
- How to create tmap labels along lines with rotated crs?
- How to access the value of a reactive output inside the browser console with jQuery?
- Convert excel column names to numbers
- How to adjust a node width in a Sankey plot in R?