Dimensionality Reduction – PCA explanation
I don't think I have a good understanding of PCA, can someone help with my confusion below please?
Take iris dataset as an example, I have 4 covariates, x1:sepal length; x2:sepal width; x3:petal length; x4:petal width. And the formula can be seen below, a1,a2,a3,a4 are the weightings for the covariates. And PCA will try to maximise the variance using different linear transformations. While also follows the rule of a1^2 + a2^2 + a3^2 + a4^2=1. I'm interested in knowing the value of a1,a2,a3,a4.
a1*x1 + a2*x2 + a3*x3 + a4*x4
I have below code on python which I think is correct?
# load libraries
from sklearn.datasets import load_iris
from sklearn.decomposition import PCA
import seaborn as sns
import pandas as pd
import numpy as np
iris = load_iris()
X = iris.data
df = pd.DataFrame(X,columns=iris.feature_names)
pca = decomposition.PCA(n_components = 4)
digits_pca_4 = pca.fit(X)
digits_pca_4.explained_variance_ratio_
And the result is
array([0.92461872, 0.05306648, 0.01710261, 0.00521218])
My question is:
Am I correct to assume that a1=sqrt(0.92), a2=sqrt(0.05), a3=sqrt(0.02), a4=sqrt(0.005)?
Second question:
And if I were to choose the linear combination of a1=a2=a3=a4=0.5, what's the variance of this compared to the variance from the PCA (I'm assuming it's less than the PCA result since PCA maximise the variance?)? How can I get the variance of when a1=a2=a3=a4=0.5 in python? And is the variance from PCA the code below?
pca.explained_variance_.sum()
To answer directly your question: no, your initial interpretation is not correct.
Explanation
The actual projection done by PCA is a matrix multiplication Y = (X - u) W where u is the mean of X (u = X.mean(axis=0)) and W is the projection matrix found by PCA: a n x p orthonormal matrix, where n is the original data dimension and p the desired output dimensions. The expression you give (a1*x1 + a2*x2 + a3*x3 + a4*x4) does not mean anything with all values being scalars. At best, it could mean the calculation of a single component, using one column j of W below as the a_k: Y[i, j] == sum(W[k, j] * (X[i, k] - u[k]) for k in range(n)).
In any case, you can inspect all the variables of the result of pca = PCA.fit(...) with vars(pca). In particular, the projection matrix described above can be found as W = pca.components_.T. The following statements can be verified:
# projection
>>> u = pca.mean_
... W = pca.components_.T
... Y = (X - u).dot(W)
... np.allclose(Y, pca.transform(X))
True
>>> np.allclose(X.mean(axis=0), u)
True
# orthonormality
>>> np.allclose(W.T.dot(W), np.eye(W.shape[1]))
True
# explained variance is the sample variation (not population variance)
# of the projection (i.e. the variance along the proj axes)
>>> np.allclose(Y.var(axis=0, ddof=1), pca. explained_variance_)
True
Graphical demo
The simplest way to understand PCA is that it is purely a rotation in n-D (after mean removal) while retaining only the first p-dimensions. The rotation is such that your data's directions of largest variance become aligned with the natural axes in the projection.
Here is a little demo code to help you visualize what's going on. Please also read the Wikipedia page on PCA.
def pca_plot(V, W, idx, ax):
# plot only first 2 dimensions of W along with axes W
colors = ['k', 'r', 'b', 'g', 'c', 'm', 'y']
u = V.mean(axis=0) # n
axes_lengths = 1.5*(V - u).dot(W).std(axis=0)
axes = W * axes_lengths # n x p
axes = axes[:2].T # p x 2
ax.set_aspect('equal')
ax.scatter(V[:, 0], V[:, 1], alpha=.2)
ax.scatter(V[idx, 0], V[idx, 1], color='r')
hlen = np.max(np.linalg.norm((V - u)[:, :2], axis=1)) / 25
for k in range(axes.shape[0]):
ax.arrow(*u[:2], *axes[k], head_width=hlen/2, head_length=hlen, fc=colors[k], ec=colors[k])
def pca_demo(X, p):
n = X.shape[1] # input dimension
pca = PCA(n_components=p).fit(X)
u = pca.mean_
v = pca.explained_variance_
W = pca.components_.T
Y = pca.transform(X)
assert np.allclose((X - u).dot(W), Y)
# plot first 2D of both input space and output space
# for visual identification: select a point that's as far as possible
# in the direction of the diagonal of the axes cube, after normalization
# Z: variance-1 projection
Z = (X - u).dot(W/np.sqrt(v))
idx = np.argmax(Z.sum(axis=1) / np.sqrt(np.linalg.norm(Z, axis=1)))
fig, ax = plt.subplots(ncols=2, figsize=(12, 6))
# input space
pca_plot(X, W, idx, ax[0])
ax[0].set_title('input data (first 2D)')
# output space
pca_plot(Y, np.eye(p), idx, ax[1])
ax[1].set_title('projection (first 2D)')
return Y, W, u, pca
Examples
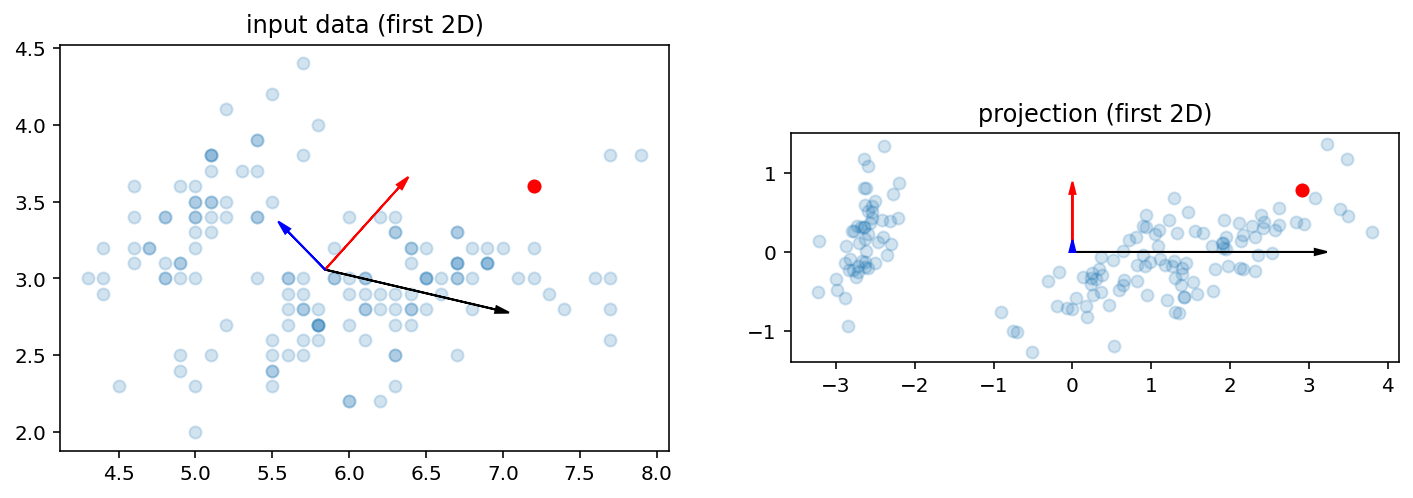
Iris data
# to better understand the shape of W, we project onto
# a space of dimension p=3
X = load_iris().data
Y, W, u, pca = pca_demo(X, 3)
Note that the projection is really just (X - u) W:
>>> np.allclose((X - u).dot(W), Y)
True
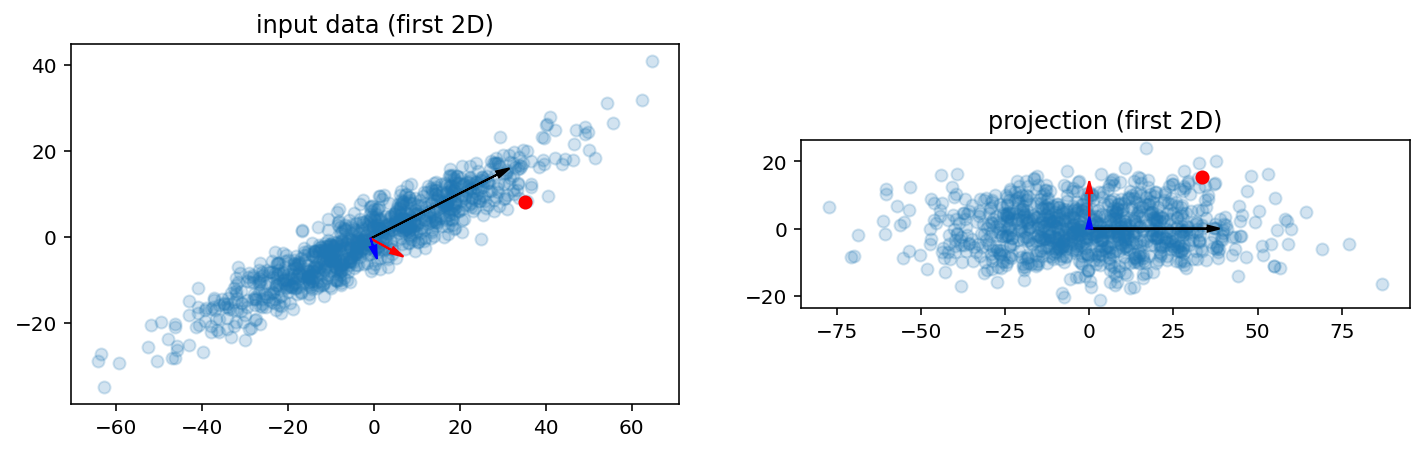
Synthetic ellipsoid data
A = np.array([
[20, 10, 7],
[-1, 3, 7],
[5, 1, 2],
])
X = np.random.normal(size=(1000, A.shape[0])).dot(A)
Y, W, u, pca = pca_demo(X, 3)
- Python, Web Scraping
- Cannot scrape xpath using Selenium
- Scrapy script not scraping items
- why does the html all have same class and sub-class with different information
- Yfinace - Getting Too Many Requests. Rate limited. Try after a while
- Requests and BeautifulSoup to get video length from YouTube
- web scraping Proplem no such element
- Python/Selenium - Scraping Into Excel Sheet
- Scraping data using Network, Fetch, Response
- Python NLTK: Bigrams trigrams fourgrams
- Discovering Poetic Form with NLTK and CMU Dict
- Python Wand: MagickReadImage returns false, but did not raise ImageMagick exception
- Executing multi-line statements in the one-line command-line
- How to get the replit link for uptime robot?
- How to install TA-Lib On vscode/windows(64bit)
- Python injector doesn't work for NewType based on tuple[str, str]
- Python conditional list joins
- Why Can't VS Code Directly Run Python unittest .py's Or Discover In Testing Tab
- Limiting amount of objects user can add in another object
- I got warning as: "Name 'x' can be undefined" and "Global variable 'x' is undefined at the module level"
- How to split data into 3 sets (train, validation and test)?
- How can I set session details in conftest.py for yield along with fixture in pytest?
- Show DataFrame as table in iPython Notebook
- How to get all possible (2^N) combinations of a list’s elements, of any length
- Python Declare variables iteratively
- Quarto Unable to Render Python
- Filter pandas dataframe with specific column names in python
- Tkinter canvas image probably being garbage collected but creating a persistent reference does not solve the problem
- Why is Pipenv not picking up my Pyenv versions?
- Why do I get lots of terminal messages about files 'first seen with mtime' when starting django runserver?