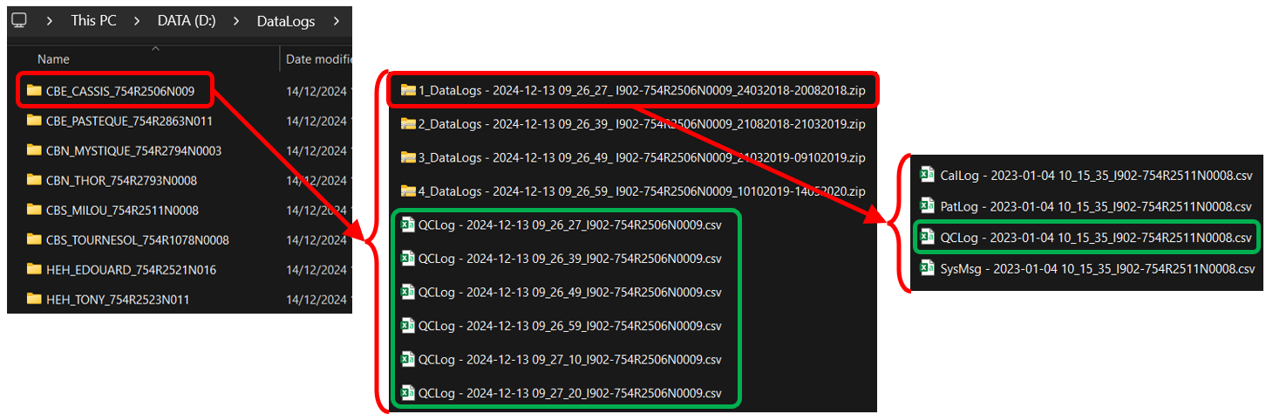
Among many subfolders, find CSVs starting with a given string and within ZIPs or not, and merge them while adding their names in a new column with R
In a folder (path = "D:/DataLogs/), I have several subfolders. Inside these subfolders, I would like to retrieve all the csv starting only with "QCLog" and merge them (rbind) into a single data.frame, while creating a first new column including the full path/name of these QCLog csv.
The two difficulties are:
- In some subfolders, there may be csv (starting with QCLog or not) directly accessible and others located in several zip files (examples within the green rectangles below).
- All CSVs have the same number of columns (n=66) and the same headers. However, some headers contain accents or symbols, almost every second column (one in two) has the exact same header name, and some columns are empty (no NAs).
Is this feasible?
Thanks for help
Solution
The following code, although a bit long, works fine and fast:
library(tidyverse)
library(data.table)
# paths
main_path <- "D:/DataLogs/" # target directory
zip_path <- "D:/DataLogs/Zip_files/" # subfolder where the zips will be unzipped
# specific string of csv file name
str_csv <- "QCLog"
# list csv
csv_nozip_list <- list.files(main_path, paste0(str_csv, ".*\\.csv"), recursive = TRUE, full.names = TRUE) # list of QCLog csv that ARE NOT within zip
zip_list <- list.files(main_path, "\\.zip", recursive = TRUE, full.names = TRUE) # list of all csv -including QCLog- that ARE within zip
unzip_files <- lapply(zip_list, unzip, exdir = zip_path) # unzip all CSV in the zip_path subfolder
csv_zip_list <- list.files(zip_path, paste0(str_csv, ".*\\.csv"), recursive = TRUE, full.names = TRUE) # list only QCLog that have been decompressed in zip_path
# read matching files while adding a new filename column
csv_nozip_read <- lapply(csv_nozip_list, function(x) {data.table(fread(x), "filename" = x)})
csv_zip_read <- lapply(csv_zip_list, function(x) {data.table(fread(x), "filename" = x)})
# rbind lists
csv_nozip_rbind <- data.table::rbindlist(csv_nozip_read)
csv_zip_rbind <- data.table::rbindlist(csv_zip_read)
# rbind selected csv
dat <- rbind(csv_nozip_rbind, csv_zip_rbind) |>
distinct() |> # if needed
relocate(filename)
- Simple question: how to add text to r chunk in rmd format?
- Issue with Label Behaviour in gganimate
- Combine census tracts with neighbor to reach a population threshold in R
- Creating a single map composed of three separate shapefiles
- Implementation of Cobb-Douglas Utility Function to calculate Receiver Operator Curve & AUC
- Overall percentages based on total count
- Pairwise dissimilarities nesting a time-series loop inside a site loop - multiple times
- Long/Lat points keep ending up in Kansas
- How to sum a column dependent on a value in another column
- Customize label for an Interaction Plot
- How to obtain RMSE out of lm result?
- Faceted mosaic plots with the same area scaling
- Unite columns with unique values
- How to print the current map while preserving data points
- How to solve error: mismatched lengths of ids and values when data has missing values in geom_line_interactive()?
- How do I transfer NAs from one dataframe to same position in a second dataframe
- Creating bar plots using ggplot, running into issues with data format
- Force initial zoom to truncate portion of the data
- R - select only factor columns of dataframe
- How to scale the units of the data & trend components of an autoplot of a multiplicative decomposition of a time series?
- Respect ratio when using ggarrange() and geom_sf()
- Changing size / aspect ratio of leaflet
- How do I use the lubridate package to calculate the number of months between two date vectors where one of the vectors has NA values?
- Adding to logos to flexdashboard
- R replace values of a column based on exact match of another data frame
- Error in grepl(pattern, df): invalid regular expression
- regular expression in R, reuse matched string in replacement
- Substract minimum value of row from each element of row in dataframe,
- R Reticulate does work with for loop but not purrr::map2
- Count all values in a correlation matrix that are above 0.8 and below -0.8