PCA after k-means clustering of multidimensional data
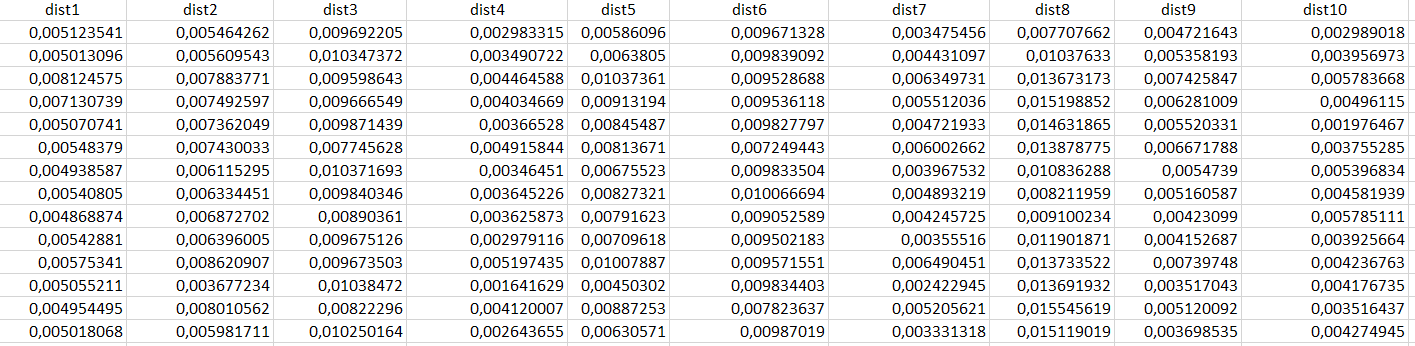
I have the following dataset with 10 variables:
I want to identify clusters with this multidimensional dataset, so I tried k-means clustering algorith with the following code:
clustering_kmeans = KMeans(n_clusters=2, precompute_distances="auto", n_jobs=-1)
data['clusters'] = clustering_kmeans.fit_predict(data)
In order to plot the result I used PCA for dimensionality reduction:
reduced_data = PCA(n_components=2).fit_transform(data)
results = pd.DataFrame(reduced_data,columns=['pca1','pca2'])
sns.scatterplot(x="pca1", y="pca2", hue=kmeans['clusters'], data=results)
plt.title('K-means Clustering with 2 dimensions')
plt.show()
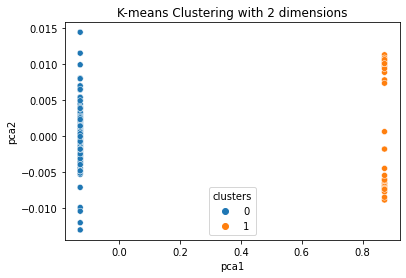
And in the end I get the following result:
So I have the following questions:
However, this PCA plot looks really weird splitting the whole dataset in two corners of the plot. Is that even correct or did I code something wrong?
Is there another algorithm for clustering multidimensional data? I look at this but I can not find an approriate algorithm for clustering multidimensional data... How do I even implement e.g. Ward hierarchical clustering in python for my dataset?
Why should I use PCA for dimensionality reduction? Can I also use t SNE? Is it better?
the problem is that you fit your PCA on your dataframe, but the dataframe contains the cluster. Column 'cluster' will probably contain most of the variation in your dataset an therefore the information in the first PC will just coincide with
data['cluster']column. Try to fit your PCA only on the distance columns:data_reduced = PCA(n_componnts=2).fit_transform(data[['dist1', 'dist2',..., dist10']]You can fit hierarchical clustering with sklearn by using:
sklearn.cluster.AgglomerativeClustering()`You can use different distance metrics and linkages like 'ward'
tSNE is used to visualize multivariate data and the goal of this technique is not clustering
- Python library for RSA public decryption
- Django: Update multiple objects attributes
- 'Unsupported Filling Mode' error while sending order from Python API to Meta Trader 5
- What does *tuple and **dict mean in Python?
- NumPy array is not JSON serializable
- Does Python have a ternary conditional operator?
- How to speed up while loop with PyAutoGui?
- How to set a single title for facetted xarray plot?
- Authenticating a user with NextCloud using oauth2 with authlib in flask fails at getting access token
- In Python, how do I iterate operation over several lists and return an updated list for each?
- Suppress InsecureRequestWarning: Unverified HTTPS request is being made in Python2.6
- How to serve Flask static files using Nginx?
- "module 'os' has no attribute 'add_dll_directory'" in aws lambda function
- Executing update statement with SQLAlchemy does not update rows, but statement works perfectly in when executed in MySQL Workbench
- Making Turtle Graphics Python game start only after main menu button has been clicked
- How do I account for a particle at rest on the ground in a gravity particle simulation?
- What's the correct way to check if an object is a typing.Generic?
- cannot perform operation: another operation is in progress in pytest
- UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
- How to annotate a generator in Python 3?
- Getting imports right using grpc
- How can I install a Python package temporarily with uv without adding it to pyproject.toml?
- How can I solve CondaHTTPError in anaconda?
- How do I read the content of a span class with python playwright
- Calculating the partial derivative of PyTorch model output with respect to neurons pre-activation
- Why am I getting a Pylance error "Object of type 'A*' is not callable" in VSCode when using generics?
- How to check if an array is multidimensional
- binning data in python with scipy/numpy
- How to rotate a page by arbitrary angle in pymupdf?
- How to make a list of checked checkboxes in pyQt